REAL-TIME 3D SINGLE-PARTICLE TRACKING AND 3D MULTI-RESOLUTION IMAGING
The interior of a cell is highly heterogeneous where molecular and nanoscale dynamics span multiple time and length scales. Furthermore, these dynamics all occur in three dimensions (3D). For example: How does a viral particle or a vaccine-delivery vehicle approach a living cell and become internalized? What are the dynamic molecule-level interactions that lead to a (un)successful entry? How does the intracellular cargo get delivered to its destination? To understand the molecule-nanoscale dynamics in the live-cell context, time must be explicitly considered. My lab recognized this problem very early on and has developed a real-time 3D single-particle tracking (RT-3DSPT) technology which enables us to follow a tagged nanoscale probe with 10 μs time resolution and ~10 nm spatial localization precision. We are also developing new ways to integrate this RT-3DSPT technology to a variety of imaging microscopy modalities as well as single-particle spectroscopy of a freely moving nanoparticle such as the measurement of single-particle 3D reorientation dynamics.
3D Real-Time Multi-Resolution Microscopy
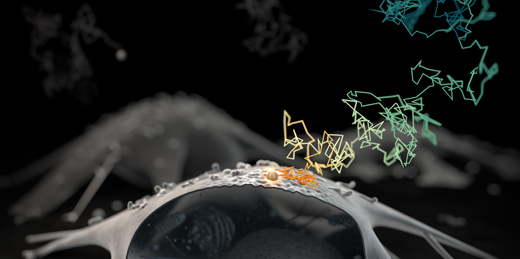
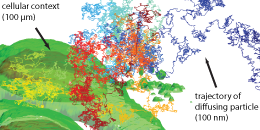
We pioneered 3D real-time multi-resolution microscopy. It is achieved by integrating, on the hardware level, the high-resolution RT-3DSPT (see below) with a lower-resolution imaging modality, taken concurrently in real time, that gives the surrounding larger environmental contexts. With this we were able to directly monitor chemical dynamics at interfaces, for example.
Links: Opt. Lett., 32, 2729-2731 (2007): conceptualization and initial realization; Nat. Nanotechnol., 9, 198-203 (2014): concurrent 3D contextual imaging realization; Faraday Discuss., 184, 359-379 (2015): technical review; J. Microbiol. Immunol. Infect., 56, 257-266 (2023): tracking individual viruses.
3D Multi-focal Volumetric Imaging at Video Rates
We have implemented a spatial light modulator (SLM) based multi-color, multi-focal microscope (MFM) to acquire 3D volume images at video rates (> 20 frames/second). We are currently using this to understand diffusion in confined environments.
Links: PLoS ONE, 15, e0230217 (2020): new approach to achieve uniform intensity; J. Opt. Soc. Am. B, 38, 2792-2798 (2021): MFM localization precision; Proc. SPIE, XXVIII, 11649OP (2021): SLM-MFM optimization algorithm; Sci. Rep., 12, 26343 (2022): multi-color MFM.
Real-Time 3D Single-Particle Tracking & Spectroscopy
We pioneered real-time 3D single-particle tracking and have since been continually furthering the concept both technically and theoretically. Most recently, we have implemented hardware-based lifetime gating to achive much improved signal-to-background ratio.
Links: J. Chem. Phys., 155, 164201 (2021): lifetime-gated RT-3DSPT.Updated: 2024.02.23.